2024 Nobel Prize in Chemistry winners American biochemist David Baker, from left, American researcher John Jumper and Demis Hassabis, CEO of DeepMind Technologies, the AI division behind Gemini.
| Photo Credit: UW Medicine/Google DeepMind via AP/AP Photo/Jeff Chiu
The story so far: The 2024 Nobel Prize for chemistry was jointly awarded to David Baker for his research in the field of computational protein design and to Demis Hassabis and John M. Jumper for their work in protein structure prediction. The Swedish Academy of Royal Sciences announced the winners on October 9.
Why are proteins important?
The chemistry prize concerns two areas in the field of protein research: design and structure.
All life (as we know it) requires proteins and all proteins are made of amino acids. While there are many types of amino acids in nature, only 20 of them in different combinations make up all the proteins in the human body and in most life-forms.
Amino acids are found in tissues — like muscles, skin, hair — that provide structural support, serve as catalysts for biochemical reactions, transport molecules like oxygen across biological membranes, control muscle contraction which lets us move and walk and makes our heart beat, and regulate cell communication that allow different functions to occur efficiently.
What is the protein-folding problem?
The arrangement of amino acids relative to one another determines which function a particular protein will serve. Understanding the 3D shapes of proteins is thus key. Scientists have spent decades trying to decode this. In 1962, University of Cambridge researchers John Kendrew and Max Perutz won the chemistry Nobel Prize for elucidating the first 3D models of haemoglobin and myoglobin (both proteins) using X-ray crystallography. In 1961, Christian Anfinsen found that a protein’s 3D structure is governed by the sequence of amino acids in the protein, and he won the 1972 chemistry prize.
One notable breakthrough arrived in 1969, when scientists found that a protein doesn’t try to bend into different shapes before settling into its final one. Instead it somehow knows the shape it needs to get to and gets there very quickly. The mysterious nature of this ‘knowledge’ of the protein is called the protein-folding problem.
By the late 2010s, scientists had worked out the structures of around 1.7 lakh proteins — a large number yet still small compared to the roughly 200 million proteins in nature. This situation changed drastically around 2018.
What is AlphaFold?
Hassabis co-founded DeepMind in 2010 and which Google acquired in 2014. Here, Hassabis, Jumper, and others built and unveiled AlphaFold in 2018. AlphaFold is a deep-learning model able to predict the structures of proteins after training on the 1.7 lakh known structures. DeepMind launched its successor AlphaFold 2 in 2021.
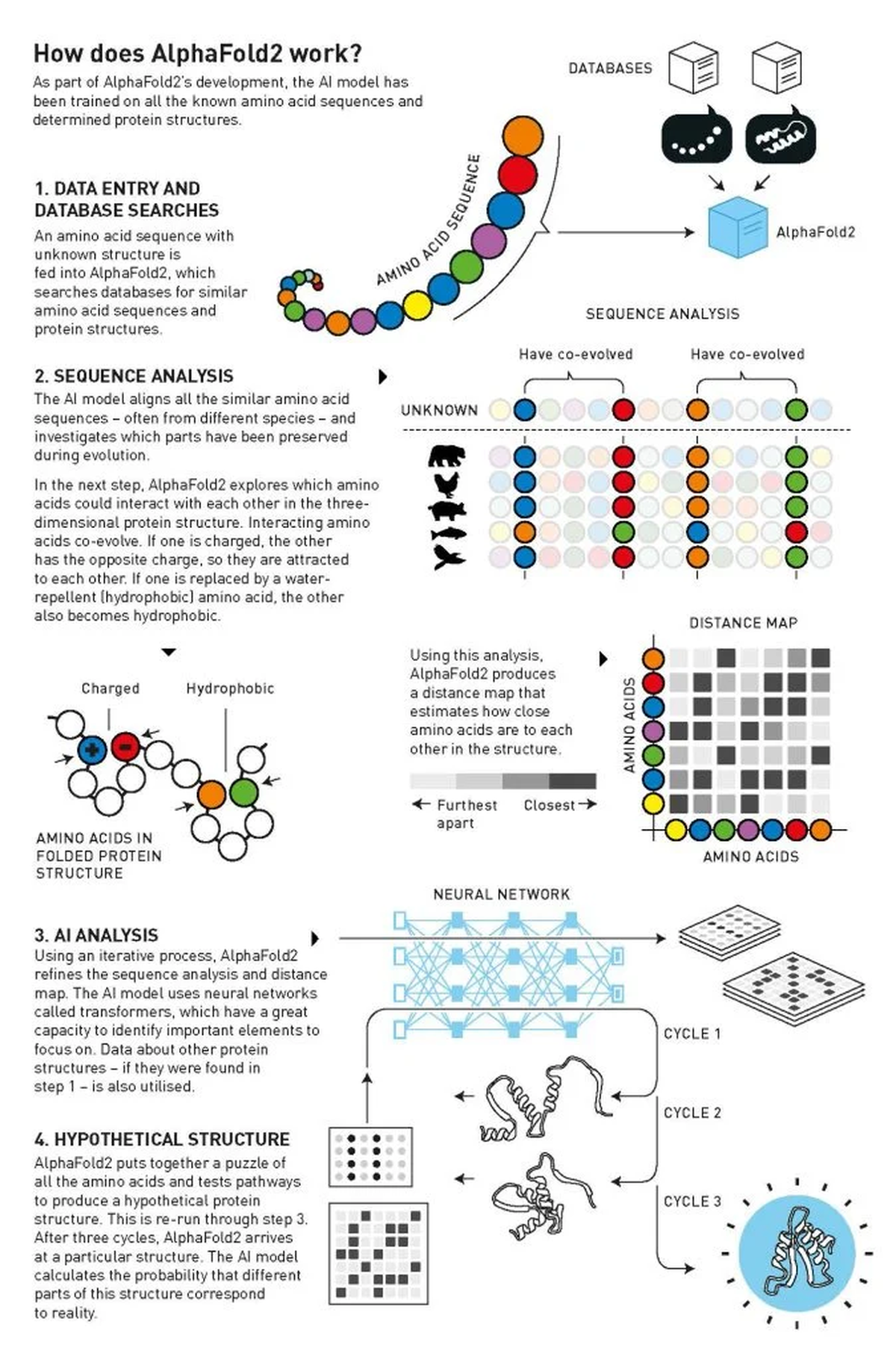
How does AlphaFold2 work?
| Photo Credit:
Johan Jarnestad/The Royal Swedish Academy of Sciences
AlphaFold is essentially a pattern-spotter on steroids. Its forté is to zero in on the most likely structure of a given protein rather than reason its way to the answer based on any precepts of structural biology.
“If the protein folding problem was set to us by God to teach us how to learn molecular interactions from first principles, we cheated,” Derek Lowe, a pharmaceutical researcher and author of a column in Science, told The Hindu in June 2024. “We haven’t learned a tremendous amount more about that. We have figured out how they usually do it, even if we don’t know why.”

Jumper led the work on AlphaFold 3, which DeepMind released in May 2024. This model is able to predict the structures of various proteins as well as how two proteins and/or a protein and another molecule might interact.
Given enough computing power, these machine-learning models are capable of deducing the 3D shapes of proteins in a matter of hours — a task that once occupied humans scientists for several months, if not years.
What is protein design?
Baker, who received the other half of this year’s chemistry Nobel Prize, developed tools that scientists use to design new proteins with specific shapes and functions. His first notable work was in 2003, when he led a team created a novel protein and determined its structure using a computer program they wrote, called ‘Rosetta’. The researchers also used X-ray crystallography to determine its structure. The outcomes of the two methods were remarkably similar.
Rosetta works by picking shorter protein fragments from a database and tying them together to predict the structure of the protein in question, together with predictive methods to optimise the sequence and the structure.
The ability to design proteins has far-reaching implications. For example, in 2022, Baker’s team developed an antiviral nasal spray to treat COVID-19. At its heart were proteins the team designed using computational methods in the laboratory. They could stick to vulnerable sites on the viral surface and target the spike protein.
Published – October 09, 2024 09:23 pm IST







